Softwares
DISPbind: disorder protein genomic binding analysis toolkit for DisP-seq
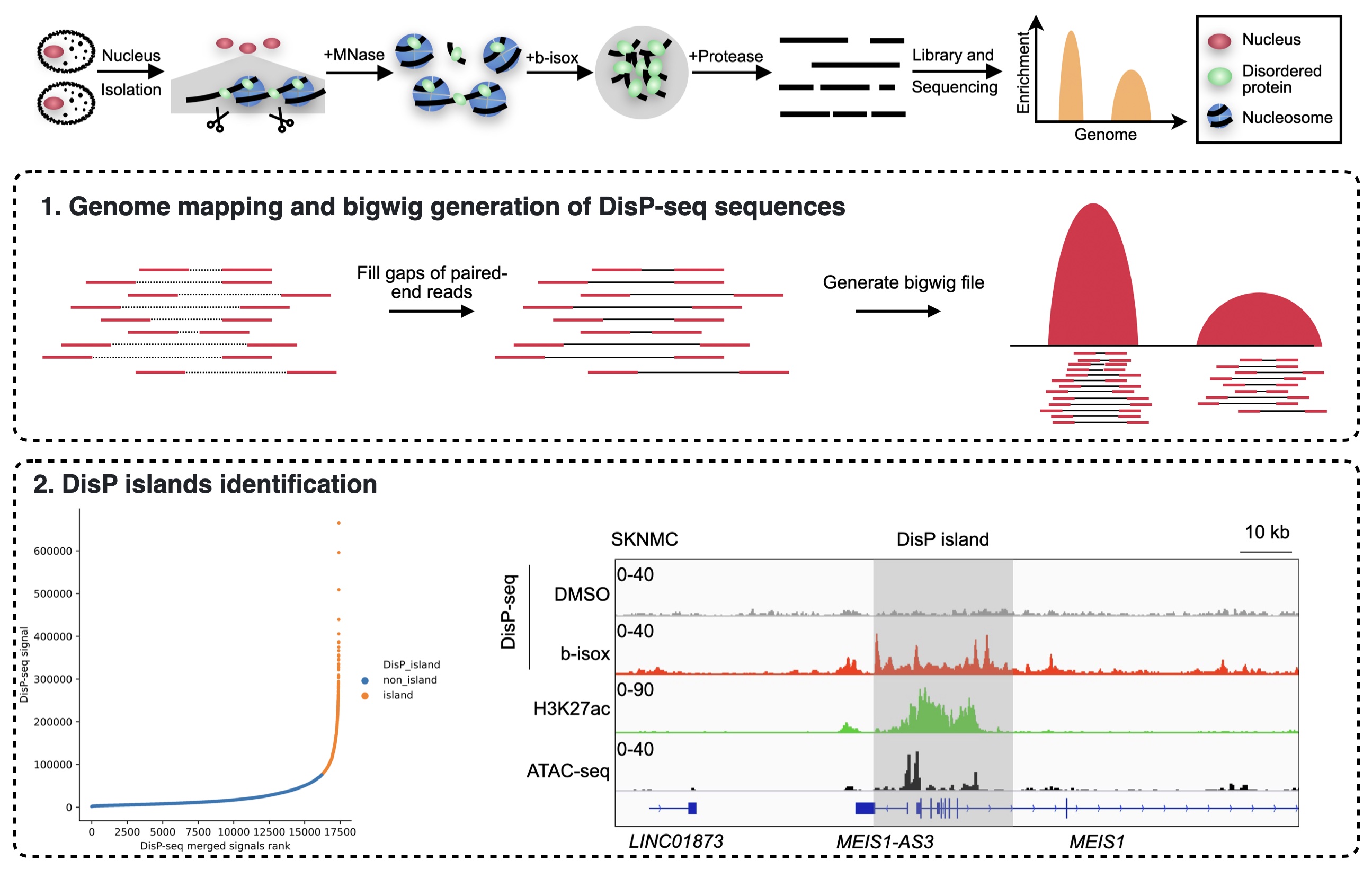
DISPbind is designed for DisP-seq datasets, providing essential functionalities such as genome mapping, bigwig generation and DisP islands identification.
Github: https://github.com/rdong08/DISPbind
Citation: Xing YH#, Dong R#, Lee L, Rengarajan S, Riggi N, Boulay G, Rivera N. M*. DisP-seq reveals the genome-wide functional organization of DNA-associated disordered proteins. Nat. Biotechnol., 2023: 1-13.
SpatialDWLS: accurate deconvolution of lower resolution spatial transcriptomic data
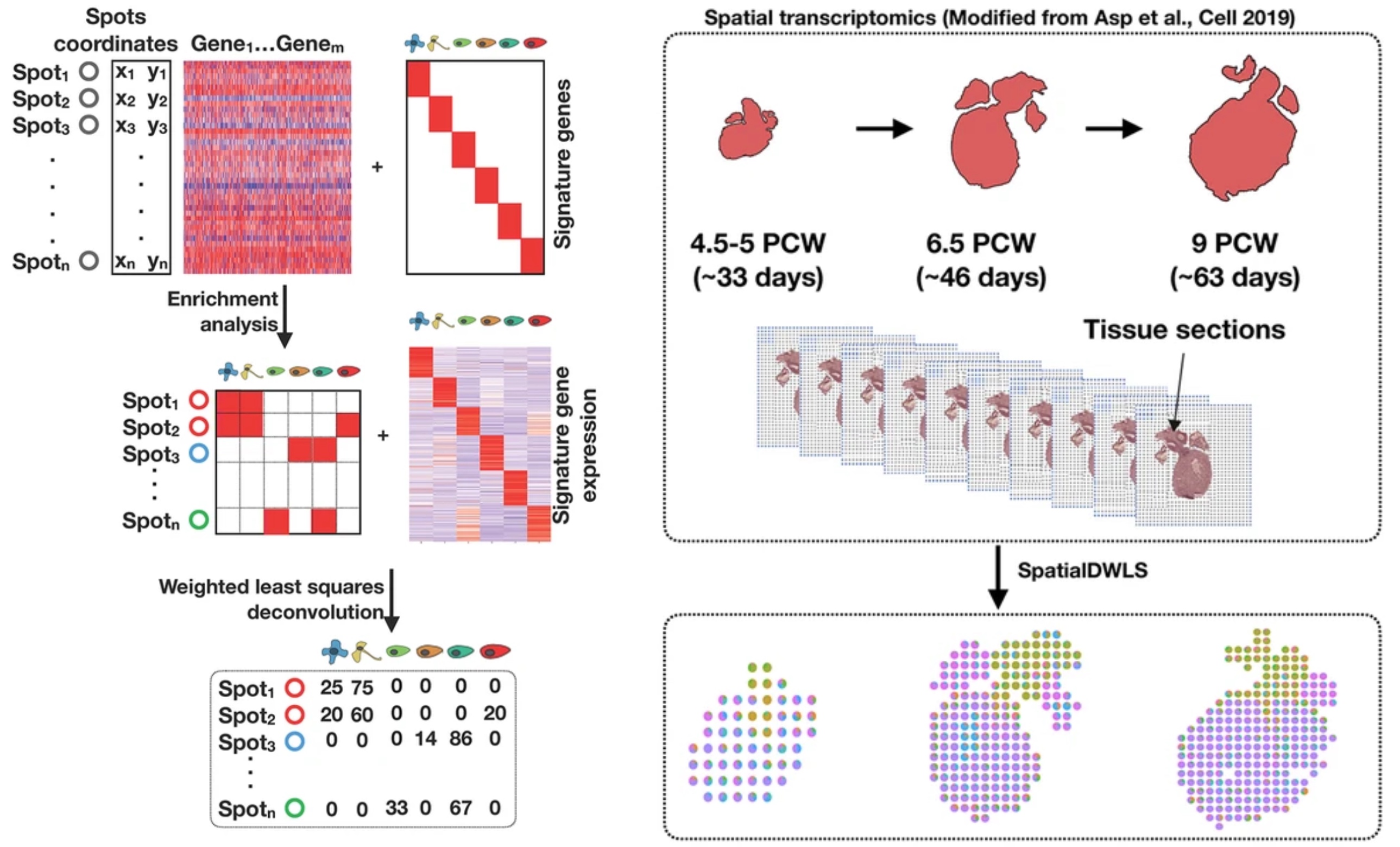
SpatialDWLS is an accurate and computationally efficient method for estimating the spatial distribution of cell types from spatial transcriptomic data. It provides a valuable enabling toolkit for investigating cell-cell interactions from various spatial transcriptomic technology platforms that do not have single-cell resolution.
Github: SpatialDWLS example dataset. The SpatialDWLS has beed implemented into Giotto.
Citation: Dong R, Yuan GC*, SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol., 2021, 22(1): 1-10.
GiniClust3: rare cell identification from large scale single cell RNA-seq datasets
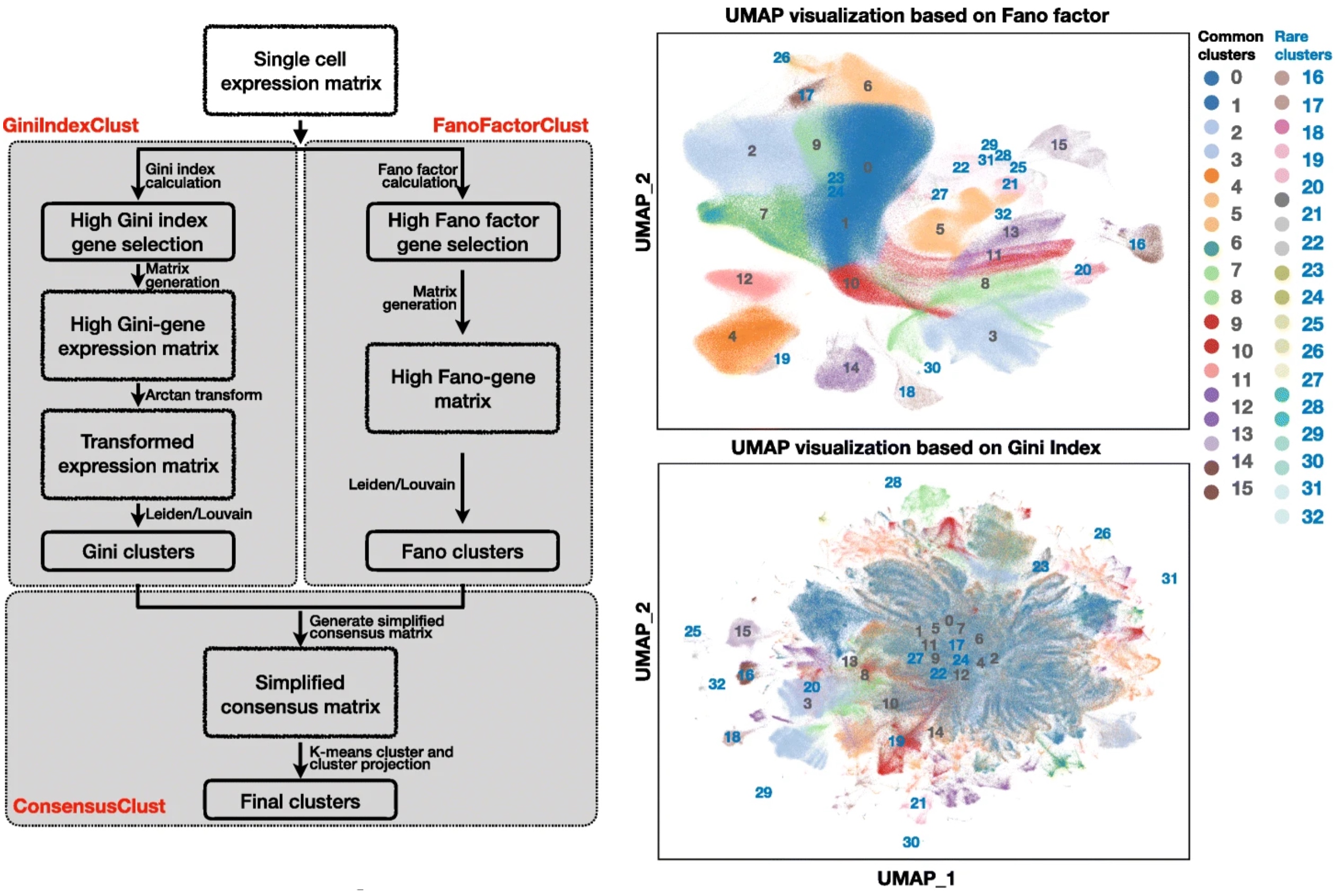
GiniClust3 is specifically tailored for large-scale scRNA-seq dataset for rare cell type identification.
Github: https://github.com/rdong08/GiniClust3
Citation: Dong R, Yuan GC*, GiniClust3: a fast and memory-efficient tool for rare cell type identification. BMC Bioinform., 2020 21: 1-7.